Project outline
Initiative
HMC, the Helmholtz Metadata Collaboration, is a network within the Helmholtz Association, aiming to foster and to improve metadata at all levels and in all scientific domaines of the Helmholtz Association.
HMC provides yearly calls for proposal of projects
which can run up to 2 years,
include at least 2 Helmholtz centers,
and are supported by up to 200.000,00 € additional funding.
In addition, HMC provides many opporutunities to get in touch
with other HMC projects in order to learn from each other
with HMC hub representatives which have a broader overview of
all HMC projects
other german, european and international projects
or join various webinars, organized by HMC.
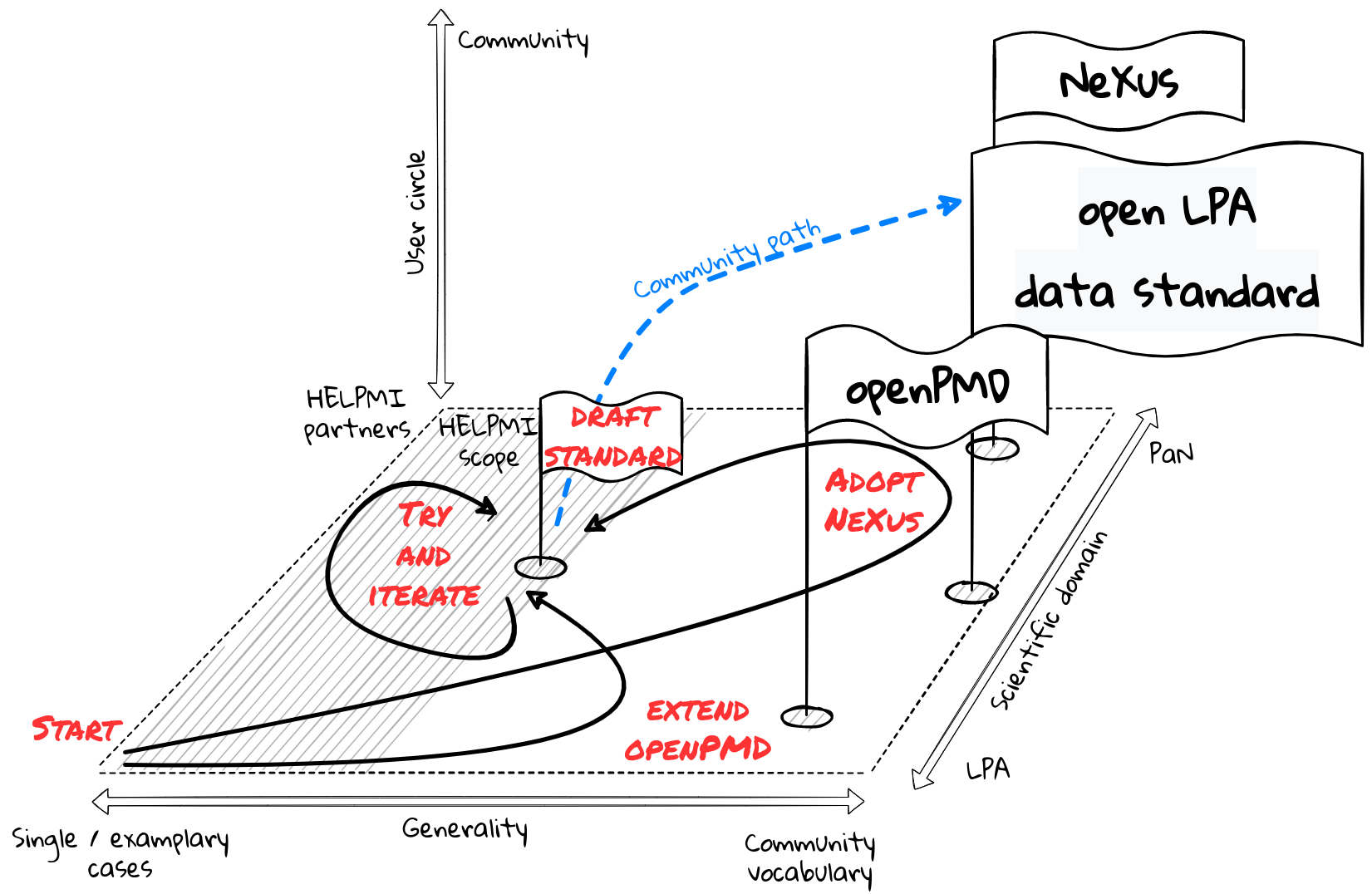
Thus, applying for HELPMI within the HMC framework provided a unique opportunity for the LPA community to start a data standard for experimental data because of the specific support and background provided from HMC. Yet, it is absolutely undisputed that the LPA community has to continue the initial efforts of HELPMI.
Glossary and Ontology
Stage 1: Glossary prototype
Any standard requires conventions. HELPMI follows a staged approach and is developing
a list of terms common in the labs of the HELPMI partners.
this list can be extended be either the partners or by the LPA community
this list can have some preliminary structure (hierarchy), but this structure can be changed or different hierarchies for the same list can be constructed as trial This list and its potential structure is to be discussed among the LPA community.
This list is to be used as a repository for keywords in data files. HELPMI will start with examplary use of terms in example data files.
Stage 2: Glossary
At a later stage, this list of terms can be curated in a human-readable and machine-readable way. There are several possibilities:
.md file on as GitHub release, to be rendered also like this web site.
GitHub allows for collaborative and documented progress
Releases can serve for unique versions
can be accessed from other codes
JSON schema on GitHub
JSON schemas are machine-actionable
in particular, tools exist which can validate whether data complies with a schema, i.e. whether metadata is complete.
Tools exist to make them human-readable
Stage 3: Ontology
Using standards of ontology description and knowledge of the relations between terms, an ontology can be built from the glossary:
other standard of term definition, machine-actionable and with tools human-readable
versioning
inter-relations
A different kind of hosting can be required, all terms have a unique identifiers. The benefit is that logical reasoning can be applied.
Instructive example: Electron Microscopy Glossary , Explain a Glossary , source repositories
Data standards
Nexus
NeXus is a family of standards, currently developed for experimental data at sychrotron radiation and neutron/muon facilities. It consists of a series of “base classes”, which can be bundled in various ways for various applications as “application definitions”. The scope can be extended by “contributed definitions”, both as additional base class or application definitions.
NeXus implements data as hdf5 files with additional attributes. A core component is the base class “NXdata” which links to data to be displayed by default, thereby enabling default views to data.
HELPMI has implemented example hdf5 files of LPA experiment and laser data with added NeXus attributes.
openPMD
The Open Standard for Particle-Mesh Data (openPMD) is a F.A.I.R. metadata standard for tabular (particle/dataframe) and structured mesh data in science and engineering. This standard has initially been concepted for – but is not restricted to – simulation codes by the computational Laser-Plasma community. Conceptually, the standard to be explored within the HELPMI project should take a position that mediates between both openPMD and NeXus by targeting laser-plasma experiments.
Taking part in the early stages of exploring such an experiments standard is an ideal condition for emphasizing the interoperability between an eventual HELPMI standard and openPMD. Conversely, HELPMI is a chance for openPMD to bridge into experiment workflows and existing standards such as NeXus.
From openPMD’s point of view, this means in a first step to inquire both the standard’s and the reference implementation’s (openPMD-api) capabilities for user-defined custom extensions, an endeavour with a wide range of applications within and outside the HELPMI project. In a second step, workflows should be examined which use these fundaments for building a form of interoperability between openPMD and HELPMI/NeXus. Possible approaches include the embedment of NeXus classes side-by-side with openPMD markup, the use of openPMD markup within newly-defined NeXus classes or the creation of openPMD layers for NeXus-defined data and vice versa.
Use cases include the comparison of experiment data against simulation data, the use of simulation tooling for experiment data or the integration into complex scientific processes that span simulation and experiment components.
The openPMD-related results of the HELPMI project will be submitted in terms of public Pull Requests on GitHub. First usable results are:
openPMD as a configuration template for experiments workflows
Support for Hard-/Softlinks in HDF5: TBD